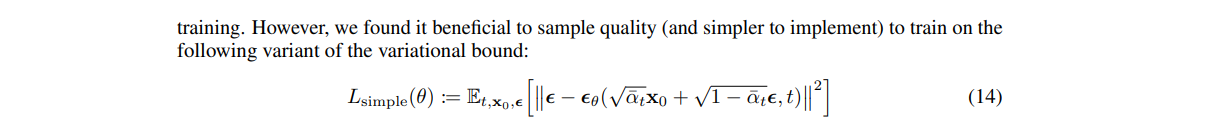The score of a probability density p is the gradient of its log-density: ∇logp(x). This object is of paramount importance in many fields, like physics, statistics, and machine learning. In particular, it is needed if one wants to sample from the reverse SDE of a diffusion model. In this note, we survey the classical technique used for learning the score of a density from its samples: score matching.
The Fisher Divergence
The L2-distance between the scores of two probability densities is often called the Fisher divergence:
fisher(ρ1∣ρ2)=∫ρ1(x)∣∇logρ1(x)−∇logρ2(x)∣2dx=EX∼ρ1[∣∇logρ1(X)−∇logρ2(X)∣2].
Since our goal is to learn ∇logp(x), it is natural to choose a parametrized family of functions sθ and to optimize θ so that the divergence
∫p(x)∣∇logp(x)−sθ(x)∣2dx
is as small as possible. However, this optimization problem is intractable, due to the presence of the explicit form of p inside the integral. This is where Score Matching techniques come into play.
The score matching trick
Let p be a smooth probability density function supported over Rd and let X be a random variable with density p. The following elementary identity is due to Hyvärinen, 2005.
Let
s: Rd→Rd be any smooth function with sufficiently fast decay at
∞, and
X∼p. Then,
E[∣∇logp(X)−s(X)∣2]=c+E[∣s(X)∣2+2∇⋅s(X)] where
c is a constant not depending on
s.
Proof. We start by expanding the square norm: ∫p(x)∣∇logp(x)−s(x)∣2dx=∫p(x)∣∇logp(x)∣2dx+∫p(x)∣s(x)∣2dx−2∫∇logp(x)⋅p(x)s(x)dx. The first term does not depend on s, it is our constant c. For the last term, we use ∇logp=∇p/p then we use the integration-by-parts formula:
2∫∇logp(x)⋅p(x)s(x)dx=2∫∇p(x)⋅s(x)dx=−2∫p(x)(∇⋅s(x))dx
and the identity is proved.
The loss we want to minimize is thus
θ∈θargminE[∣∇logp(X)−sθ(X)∣2]=θargminE[∣sθ(X)∣2+2∇⋅(sθ(X))].
Practically, learning the score of a density p from samples x1,…,xn is done by minimizing the empirical version of the right-hand side of (3):
ℓ(θ)=n1i=1∑n∣sθ(xi)∣2+2∇⋅(sθ(xi)).
This looks computable… except it's not ideal. Suppose we perform a gradient descent on θ to find the optimal θ. Then at each gradient descent step, we need to evaluate sθ as well as its divergence; and then compute the gradient in θ of the divergence in x, in other words to compute ∇θ∇x⋅sθ. In high dimension, this can be too costly.
Fortunately, there is another way to perform score matching when pt is the distribution of a random variable corrupted with additive noise, as in our setting. The following result is due to Vincent, 2010.
Denoising Score Matching Objective
Let s:Rd→Rd be a smooth function. Let X be a random variable with density p, and let ε be an independent random variable with density g. We call pnoisy the distribution of Y=X+ε. Then, E[∣∇logpnoisy(Y)−s(Y)∣2]=c+E[∣∇logg(ε)−s(Y)∣2] where c is a constant not depending on s.
Proof. Note that pnoisy=p∗g. By expanding the square, we see that E[∣∇logpnoisy(X+ε)−s(X+ε)∣2] is equal to
c+∫pnoisy(x)∣s(x)∣2dx−2∫∇pnoisy(x)⋅s(x)dx.
Now by definition, pnoisy(x)=∫p(y)g(x−y)dy, hence ∇pnoisy(x)=∫p(y)∇g(x−y)dy, and the last term above is equal to −2∫∫p(y)∇g(x−y)⋅s(x)dxdy=−2∫∫p(y)g(x−y)∇logg(x−y)⋅s(x)dydx=−2E[∇logg(ε)⋅s(X+ε)]. But then, upon adding and subtracting the term E[∣∇logg(ε)∣2] which does not depend on s, we get another constant c′ such that
E[∣∇logpnoisy(X)−s(X)∣2]=c′+E[∣∇logg(ε)−s(X+ε)∣2].When g is a centered Gaussian density, ∇logg(ε)=(−ε/Var(ε))g(ε), hence everything is known in the formula, except for the E but this can be replaced by an empirical average over the training dataset.
Although this way of presenting score matching was popular in 2020-2022, it happens to be an avatar of a deeper, clearer result called Tweedie's formula. Although there is strictly nothing more in Tweedie's formula than in DNS, the presentation and derivation of the main formulas tells a richer story, and is now considered as a solid grounding for presenting score-matching techniques in diffusions.
Herbert Robbins is often credited with the first discovery of Tweedie's formula in 1956 in the context of exponential (Poisson) distributions; a paper by Koichi Miyasawa (1961) was later rediscovered, with the first true appearance of the formula in a general, bayesian context. Miyazawa also computed second-order variants. Maurice Tweedie extended Robbin's formula, and Bradley Efron popularized it in his excellent paper on selection bias.
Tweedie's formulas
Let X be a random variable on Rd with density p, and let ε be an independent random variable with distribution N(0,σ2Id). We note pσ the distribution of Y=X+ε. Then, ∇logpσ(y)=−E[σ2ε∣∣∣∣Y]∇2logpσ(y)=−σ2Id+Cov(σ2ε∣∣∣∣Y). These expressions are equivalent to the noise prediction formulas, E[ε∣Y]=−σ2∇logpσ(Y)Cov(ε∣Y)=σ2Id+σ4∇2logpσ(Y). and to the data prediction or denoising formulas, E[X∣Y]=Y+σ2∇lnpσ(Y)Cov(X∣Y)=σ2Id+σ4∇2logpσ(Y).
Finally, the optimal denoising error (the Minimum Mean Squared Error) is given by MMSE=E[∣X−E[X∣Y]∣2]=Tr(Cov(X∣Y))=σ2d+σ4Tr(∇2logpσ(Y)).
The classical "Tweedie formula" is the first one in (14). The quantity E[X∣Y] is the best guess of the original sample X given its corrupted version Y, hence the term "denoising formula".
Proof. We simply compute ∇logpσ and ∇2logpσ by differentiating under the integral sign. Indeed,
∇logpσ(y)=pσ(y)∫∇ylogp(x)g(y−x)dx.
Since ∇g(z)=−z/σ2g(z), the expression above is equal to −∫σ2y−xpσ(y)p(x)g(y−x)dx. The joint distribution of (X,Y) is p(x)g(y−x) and the conditional density of X given Y=y is p(x∣y)=p(x)g(y−x)/pσ(y), hence the expression above is equal to −E[ε/σ2∣Y] as requested.
Now, by differentiating logpσ twice, we get
∇2logpσ(y)=pσ(y)∫∇2p(x)g(y−x)dx−(∇logpσ(y))(∇logpσ(y))⊤.
Since ∇2g(z)=−(σ−2Id−σ−4zz⊤)g(z), we get
∇2logpσ(y)=−pσ(y)∫[σ−2Id−σ−4(y−x)(y−x)⊤]p(x)g(y−x)dx−E[σ2ε∣Y]E[σ2ε∣Y]⊤.
This is exactly −σ−2Id+σ−4(E[εε⊤∣Y]−E[ε∣Y]E[ε∣Y]⊤), or −σ−2Id+Cov(ε/σ2∣Y).
The noise prediction formulas (13) directly follow from (12). For the denoising formulas, we only have to note that since Y=X+ε, we have E[X∣Y]=Y−E[ε∣Y] and Cov(X∣Y)=Cov(ε∣Y).
Since Y is centered around X, the classical ("frequentist") estimate for X given Y is Y. Tweedie's formula corrects this estimate by adding a term accounting for the fact that X is itself random: if Y lands in a region where X is unlikely to live in, then it is probable that the noise is responsible for this, and the estimate for X needs to be adjusted.
Now, what's the link between this and Denoising Score Matching ? The conditional expectation of any X given Z minimizes the L2-distance, in the sense that E[X∣Z]=f(Z) where f minimizes E[∣f(Z)−X∣2]. Formula (14) says that ∇lnpσ(y), the quantity we need to estimate, is nothing but the « best denoiser » up to a scaling −σ2:
∇lnpσ∈argminE[∣f(X+ε)−(−ε/σ2)∣2].
I find this result to give some intuition on score matching and how to parametrize the network.
If s(x) is « noise predictor » trained with the objective E[∣s(X+ε)−ε∣2], then −s(x)/σ2 is a proxy for ∇lnpnoisy. This is called « noise prediction » training.
If s(x) had rather been trained on a pure denoising objective E[∣s(X+ε)−X∣2], then x−s(x) is a noise predictor for X, hence σ−2(s(x)−x) is a proxy for ∇lnpnoisy. This is called « data prediction » training, or simply denoising training.
Both formulations are found in the litterature, as well as affine mixes of both.
Let us apply this to our diffusion setting where pt is the density of αtX0+εt where εt∼N(0,σˉt2), hence in this case g(x)=(2πσˉt2)−d/2e−∣x∣2/2σˉt2 and ∇logg(x)=−x/σˉt2.
Data prediction model
The « data prediction » (or denoising) parametrization of the neural network would minimize the objective
∫0Tw(t)E[∣sθ(t,Xt)−αtX0∣2]dt.
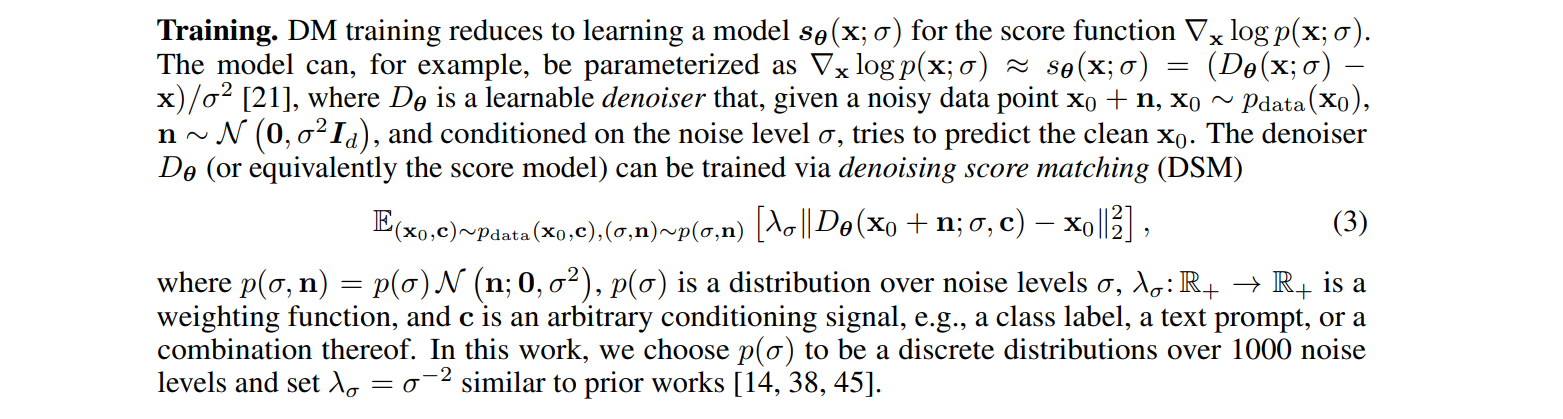
This is the loss that was used to train most (but not all) diffusion models, up to the weighting w. The SD1.5 and SDXL models use this formulation, hence if you have to use their U-NET you need to keep in mind that it's a denoiser. Here is the extract from the SDXL tech report:
Noise prediction model
Alternatively, the « noise prediction » parametrization of the neural network would minimize the objective
∫0Tw(t)E[∣sθ(t,Xt)−ε∣2]dt
Up to the constants and the choice of the drift μt and variance wt, this is exactly the loss function (14) from the early DDPM paper: 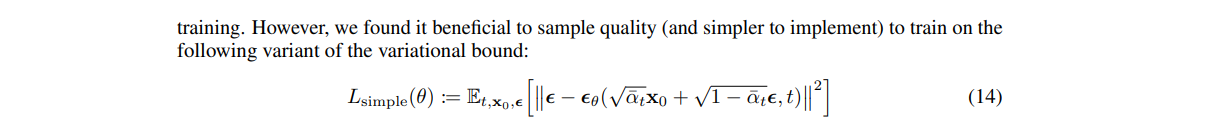
Sampling
Finally, one needs to plug these expressions back into the reverse SDE or ODE. If you want to play with pretrained models, you have to pay attention to how the model was trained: is it a data predictor or a denoiser? Based on the answer, one can go back to the score. Tweedie's formulas say that
∇logpσ(y)=σ2data predictor−y
while on the other hand
∇logpσ(y)=−σ2noise predictor.
We are now equipped with learning ∇lnpt when pt is the distribution of something like αtX0+σtε. The diffusion models we presented earlier provide such distributions given the drift and diffusion coefficients, μt and σt. But in general, we should be able to directly choose α and σ without relying on the (intricate) maths behind the Fokker-Planck equation. This is where the flow matching formulation enters the game.
Hyvärinen's paper on Score Matching.
Efron's paper on Tweedie's formula - a gem in statistics.
I didnt find a free version of Miyazawa's paper, An empirical Bayes estimator of the mean of a normal population published in Bull. Inst. Internat. Statist., 38:181–188, 1961.